xGen™ cfDNA & FFPE DNA Library Preparation Kit
High library complexity from low quality samples
The xGen cfDNA & FFPE DNA Library Preparation Kit empowers you with highly complex variant identification from degraded and low-input research samples.
xGen NGS—made for cfDNA & FFPE DNA library preparation.
Ordering
- Get high conversion rates compared to TA-ligation-based methods with novel ligase and highly modified adapters.
- Identify variants at ≤1% variant allele frequency (VAF).
- Get data from even highly degraded research samples.
Transform Your NGS Workflow with Automation
Looking to streamline your NGS workflows? Discover how automation can enhance efficiency and consistency in your lab with our NGS Automation solutions.
For customers using legacy xGen cfDNA & FFPE DNA Library Prep MC Kits, these products can be ordered here.
Australian Genome Research Facility Ltd
We've found the xGen 2x HiFi PCR Mix to be a high performing enzyme, demonstrated superior GC-bias and yielding nearly 2x more library than its competitors using the same number of cycles. This means that we can reduce the number of PCR cycles needed to achieve high yields from low inputs and reduce PCR duplicates, leading to an overall improvement in our IDT workflows.
Product details
Applications
The xGen cfDNA & FFPE DNA Library Prep MC Kit v2 produces next generation sequencing (NGS) libraries suitable for many research applications that use degraded samples, including:
- Low-frequency, somatic variant identification of SNPs
- Insertions/deletions (indels)
- Identification of inherited germline SNPs and indels
- Whole genome sequencing (WGS)
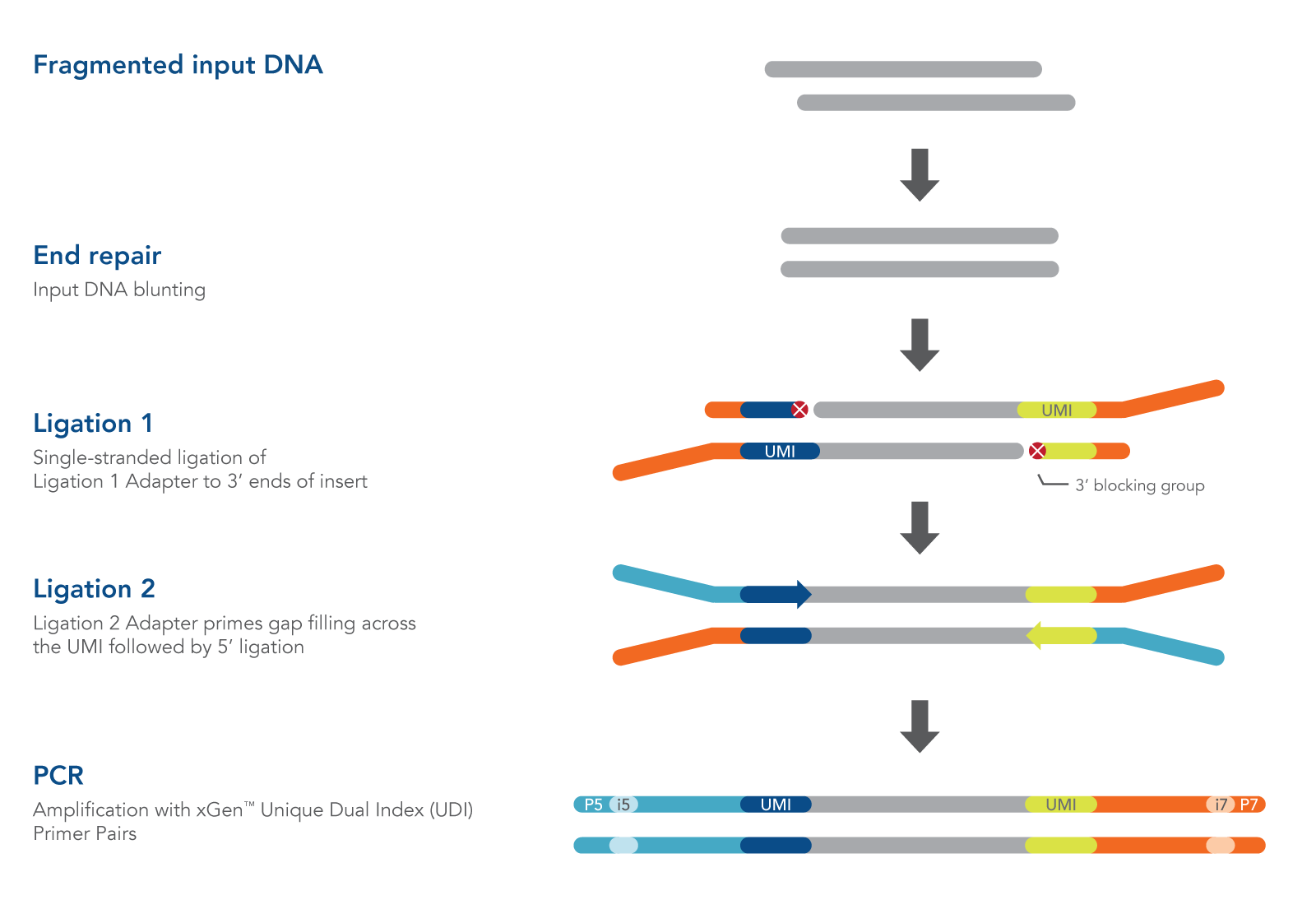
The xGen cfDNA & FFPE DNA Library Prep protocol takes about 4 hours and includes only four major steps, minimizing sample handling (Figure 1):
- End repair. The End Repair Enzyme Mix converts cfDNA, or sheared, input DNA such as FFPE DNA into blunt-ended DNA that is ready for ligation.
- Ligation 1. The Ligation 1 Enzyme catalyzes the single-stranded addition of the Ligation 1 Adapter to only the 3′ end of the insert. This novel enzyme is unable to ligate inserts together, minimizing the formation of chimeras. The 3′ end of the Ligation 1 Adapter also contains a blocking group to prevent adapter-dimer formation.
- Ligation 2. The Ligation 2 Adapter acts as a primer to gap-fill the bases complementary to the Ligation 1 Adapter, followed by ligation to the 5′ end of the DNA insert to create a double-stranded product.
- PCR amplification. The xGen 2x HiFi PCR Mix is included to perform indexing PCR (primers sold separately) for Illumina® sequencing.
Figure 1. Workflow for the xGen cfDNA & FFPE DNA Library Prep v2 MC Kit. In an initial step, end repair enzymes convert cfDNA, or sheared, input DNA into blunt-ended DNA ready for ligation. Then, a Ligation 1 Enzyme catalyzes the single-stranded addition of a Ligation 1 Adapter to the 3′ end of the insert. This novel enzyme is unable to ligate inserts together which minimizes chimera formation. The 3′ end of the Ligation 1 Adapter also contains a blocking group to prevent adapter-dimer formation. The Ligation 2 Adapter acts as a primer to gap-fill bases complementary to the Ligation 1 Adapter, followed by ligation to the 5′ end of the DNA insert which creates a double-stranded product. In a final step, PCR with the IDT 2x HiFi PCR mix incorporates sample index sequences for sequencing on Illumina® platforms.
Technical Details
The xGen cfDNA & FFPE DNA Library Prep v2 MC Kit includes all the reagents required for End Repair, Ligation 1 and Ligation 2 reactions, and PCR.
Table 1. Specifications, additional reagents, and equipment.
| Feature | Details |
|---|---|
| Sample types | High-quality DNA, cfDNA, DNA from FFPE samples |
| Input range | 1–250 ng |
| Adapters | Included |
| Indexing primers (not included) | xGen UDI Primers* |
| PCR amplification reagents | xGen 2x HiFi PCR Mix, included |
| Compatible sequencing platforms | Illumina® sequencing instruments |
| Compatible hybrid capture blockers | xGen Universal Blockers—TS Mix |
*Contact us for immediate assistance in ordering other indexing designs or configurations.
Complete workflow for hybridization capture research experiments
The xGen cfDNA & FFPE DNA Library Prep v2 MC Kit was designed to work seamlessly with xGen hybridization capture probes and reagents (Figure 2). Whether your project requires whole exome sequencing or custom panels, IDT has the capture solutions for you.
Product data
Comprehensive conversion and error correction enables ultra-low variant identification with cell-free DNA
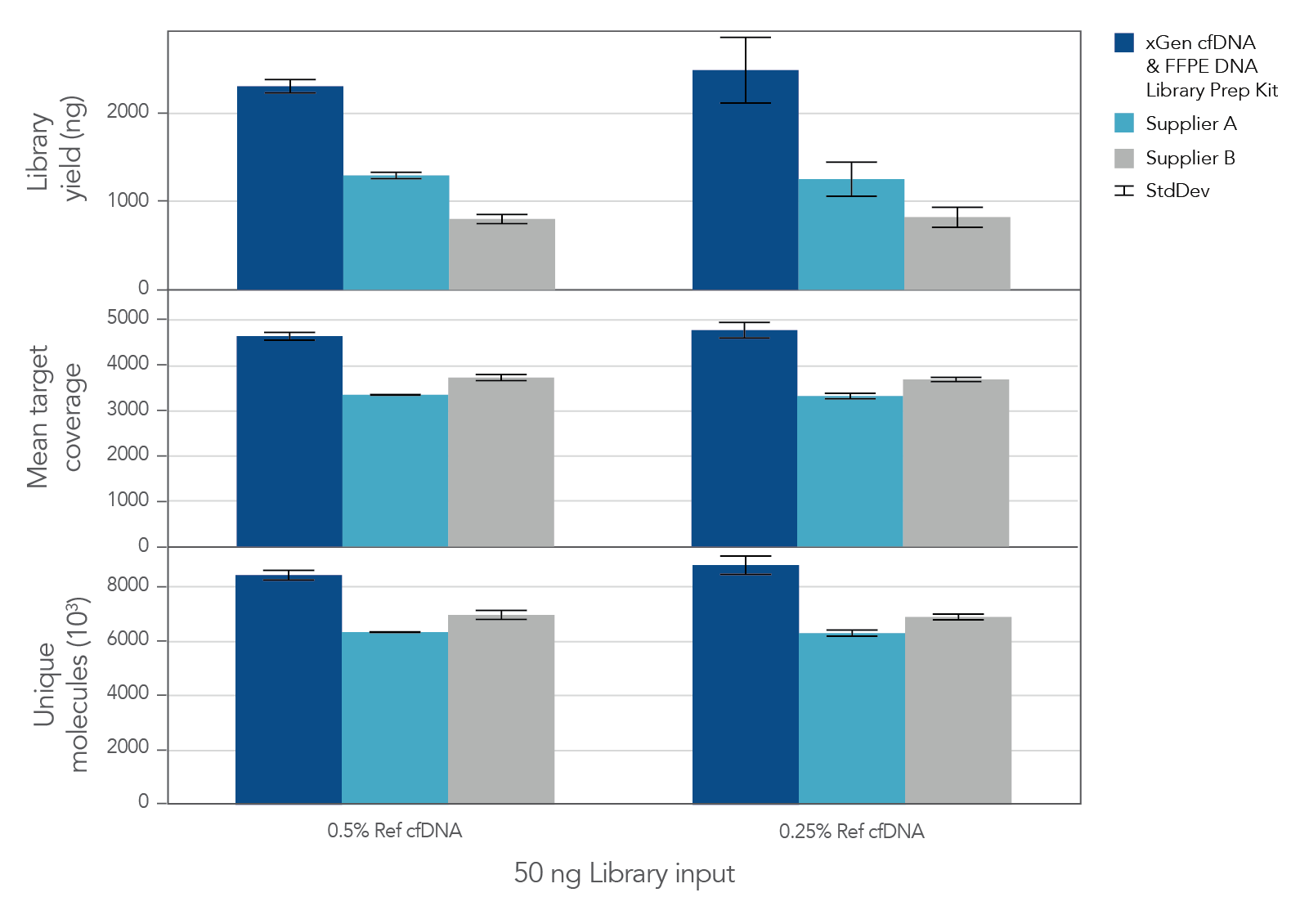
The unique, single-stranded ligation strategy of the IDT xGen cfDNA & FFPE DNA Library Prep v2 MC Kit and workflow delivers high conversion of input DNA molecules to sequencing data. This high conversion rate is critical for identification of ultra-low frequency variants, which is common in the analysis of cell-free DNA (cfDNA). A higher conversion rate translates to more complexity and coverage than other DNA library prep kits for cfDNA (Figure 3). In addition, the xGen cfDNA & FFPE DNA Library Prep Kit includes adapters that contain unique molecular identifiers (UMIs), which enable bioinformatic error correction. Combining higher complexity and coverage with stringent error correction better enables the identification of ultra-low frequency variants (Table 2).
Figure 3. xGen cfDNA &FFPE DNA Library Prep v2 MC Kit delivers higher yields, complexity, and coverage. Libraries were prepared in triplicate according to the manufacturer’s instructions with 50 ng of Horizon cfDNA reference standard and 7 cycles of PCR. Following quantification, libraries were captured with a custom 61 kb (target space) xGen Hyb Panel using the xGen Hybridization and Wash Kit. Captured libraries were pooled and sequenced on a NextSeq™ 500 (Illumina) using a high output 300 cycle kit and the manufacturer's protocol. After subsampling to 85M total reads, coverage and complexity were calculated.
Table 2. xGen cfDNA & FFPE DNA Library Prep Kit identifies low frequency variants in NGS reference samples
| Mutation | Expected VAF | xGen cfDNA & FFPE DNA Library Prep v2 MC Kit | Library Kit A | Library Kit B |
|---|---|---|---|---|
| EGFR: L858R | 0.25% | 0.13 (3/3) | 0.21 (3/3) | 0.21 (3/3) |
| EGFR: E746-A750 | 0.25% | 0.11 (3/3) | 0.19 (3/3) | 0.12 (3/3) |
| EGFR: T790M | 0.25% | 0.29 (3/3) | 0.36 (3/3) | 0.12 (3/3) |
| KRAS: G12D | 0.32% | 0.33 (3/3) | 0.36 (3/3) | 0.33 (3/3) |
| NRAS: Q61K | 0.32% | 0.23 (3/3) | 0.31 (2/3) | 0.22 (3/3) |
| NRAS: A59T | 0.32% | 0.17 (3/3) | 0.43 (2/3) | 0.22 (3/3) |
| PIK3CA:E545K | 0.32% | 0.16 (3/3) | 0.11 (3/3) | 0.36 (3/3) |
Table 2. Libraries were prepared in triplicate from 50 ng input Horizon cfDNA reference standards using the xGen cfDNA &FFPE DNA Library Prep v2 MC Kit in addition to two other commercially available library prep kits. Libraries were then captured with a custom 180 kb (target space) xGen Hyb Panel targeting 7 identified SNPs using the using the xGen Hybridization and Wash v2 Reagents and Beads. Captured libraries were pooled and sequenced on a NextSeq 500 (Illumina), using a high output 300 cycle kit and the manufacturer's protocol. After subsampling to 85M total reads, the average variant allele frequency for each of the targeted mutations was calculated for each library prep kits using VarDict.
Higher coverage and complexity deliver reliable variant and indel identification in FFPE samples
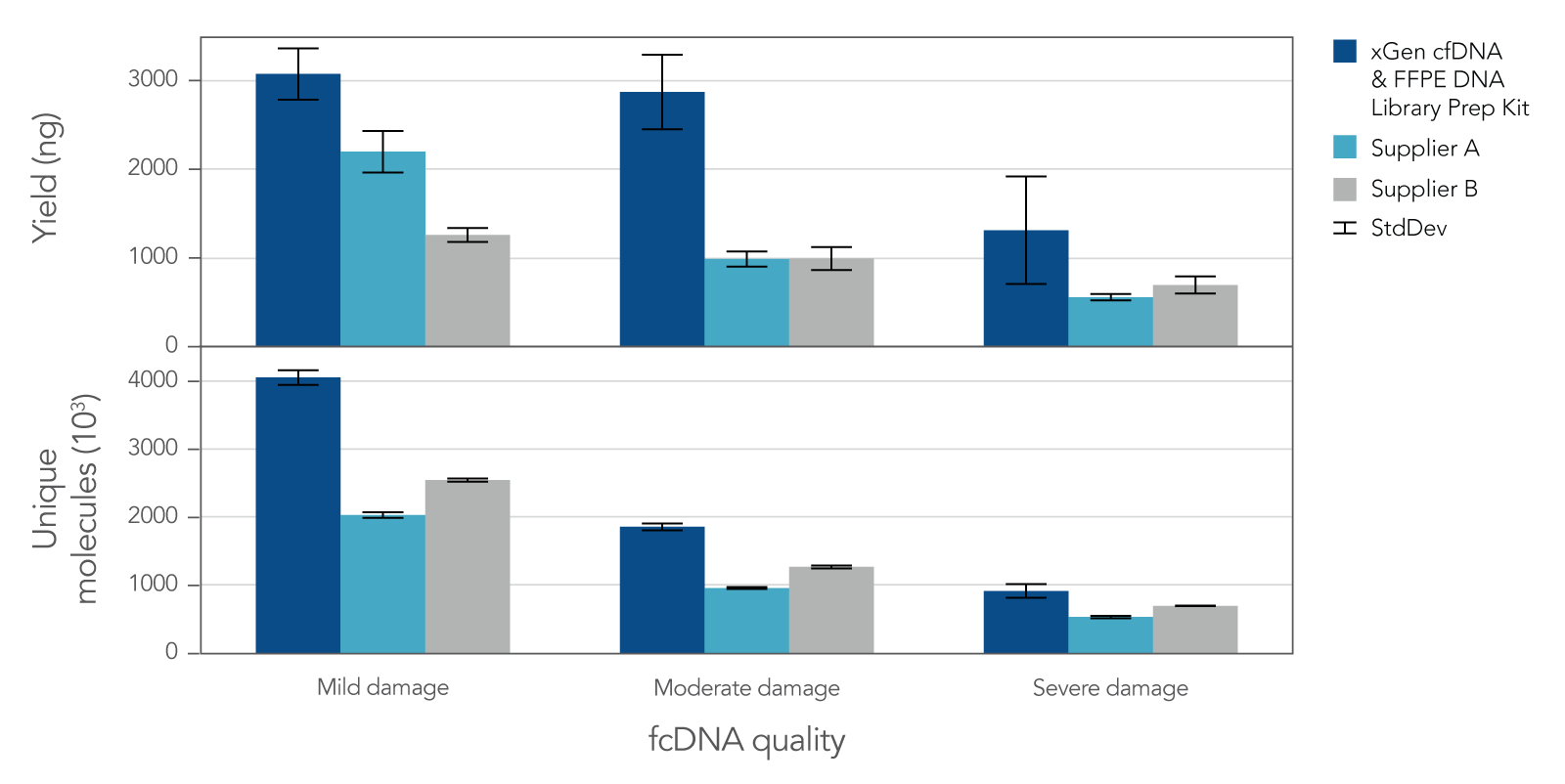
Research analysis of FFPE samples has its own unique challenges, including difficulties in generating libraries from samples of variable quality or with low inputs. The xGen cfDNA & FFPE DNA Library Prep v2 MC Kit leverages high conversion rates to achieve high library yields from low inputs of even severely damaged FFPE research samples (Figure 4). Higher conversion rates and yields translate to higher library complexity (Figure 4), which can increase confidence in variant calling. With severely damaged FFPE samples, the xGen cfDNA & FFPE DNA Library Prep v2 Kit delivers SNP and indel identification across a range of inputs (Table 3).
Figure 4. Library yield and complexity from varying qualities of Formalin Compromised DNA (fcDNA) reference standards. Libraries (n = 4 each) were prepared in triplicate by three commercial library prep kits following each of the manufacturer’s protocols with 25ng total input. Following 10 cycles of PCR, libraries were quantified using a Qubit™ dsDNA HS Kit. Then, 500 ng of each library were captured using a custom 180 kb (target space) xGen Hyb Panel with the xGen Hybridization and Wash v2 Reagents and Beads, following the xGen Hybridization and Wash Kit v2 protocol with an overnight hybridization and 13 cycles of PCR. The captured libraries were then pooled equimolar and sequenced on a NextSeq 500 (Illumina), using a high output 300 cycle kit and the manufacturer's protocol. Samples were subsampled to 8 million paired end reads and the number of unique molecules (HS library size) was determined with Picard.
Table 3. Observed Average Mutation Allelic Frequency of Severely Degraded FFPE DNA Library Input
| Variant | Expected VAF | 25ng | 50ng | 100ng | 250ng |
|---|---|---|---|---|---|
| EGFR: G719S | 24.5 | 23.9 (3/3) | 22.9 (3/3) | 22.1 (3/3) | 23.0 (3/3) |
| PIK3CA:H1047R | 17.5 | 16.9 (3/3) | 18.6 (3/3) | 17.5 (3/3) | 18.0 (3/3) |
| KRAS: G13D | 15.0 | 12.1 (3/3) | 13.2 (3/3) | 12.1 (3/3) | 12.0 (3/3) |
| NRAS: Q61K | 12.5 | 7.9 (3/3) | 11.3 (3/3) | 8.7 (3/3) | 8.9 (3/3) |
| BRAF: V600E | 10.5 | 11.2 (3/3) | 11.1 (3/3) | 11.0 (3/3) | 10.6 (3/3) |
| KIT: D816V | 10.0 | 8.0 (3/3) | 8.4 (3/3) | 7.7 (3/3) | 7.7 (3/3) |
| PIK3CA: E545K | 9.0 | 6.0 (3/3) | 7.0 (3/3) | 6.1 (3/3) | 6.3 (3/3) |
| KRAS: G12D | 6.0 | 5.7 (3/3) | 5.2 (3/3) | 6.3 (3/3) | 5.8 (3/3) |
| EGFR: L858R | 3.0 | 3.2 (3/3) | 2.9 (3/3) | 2.8 (3/3) | 2.5 (3/3) |
| EGFR: E746-A750 | 2.0 | 0.8 (3/3) | 0.5 (3/3) | 0.8 (3/3) | 0.5 (3/3) |
| EGFR: T790M | 1.0 | 0.9 (3/3) | 0.7 (3/3) | 0.8 (3/3) | 0.9 (3/3) |
Table 3. xGen cfDNA & FFPE DNA Library Prep v2 MC Kit enabled the mutation detection in severely degraded FFPE reference sample at varying library input. Each sample was captured with a custom 180 kb (target space) xGen Hyb panel targeting the verified mutations, using xGen Hybridization and Wash v2 Reagents and Beads, following the xGen Hybridization and Wash Kit v2 Protocol and sequenced to an average depth of 30 million read pairs on a NextSeq 500 (Illumina) using a high-output 300 cycle kit and the manufacturer's protocol. The average variant allele frequency (VAF) was calculated for each of the 11 mutations across all three replicates using VarDict, with a minimum variant allele depth of 3 single-stranded consensus reads. The number of replicates with a response is shown in parentheses.
Uniform GC coverage across the human genome
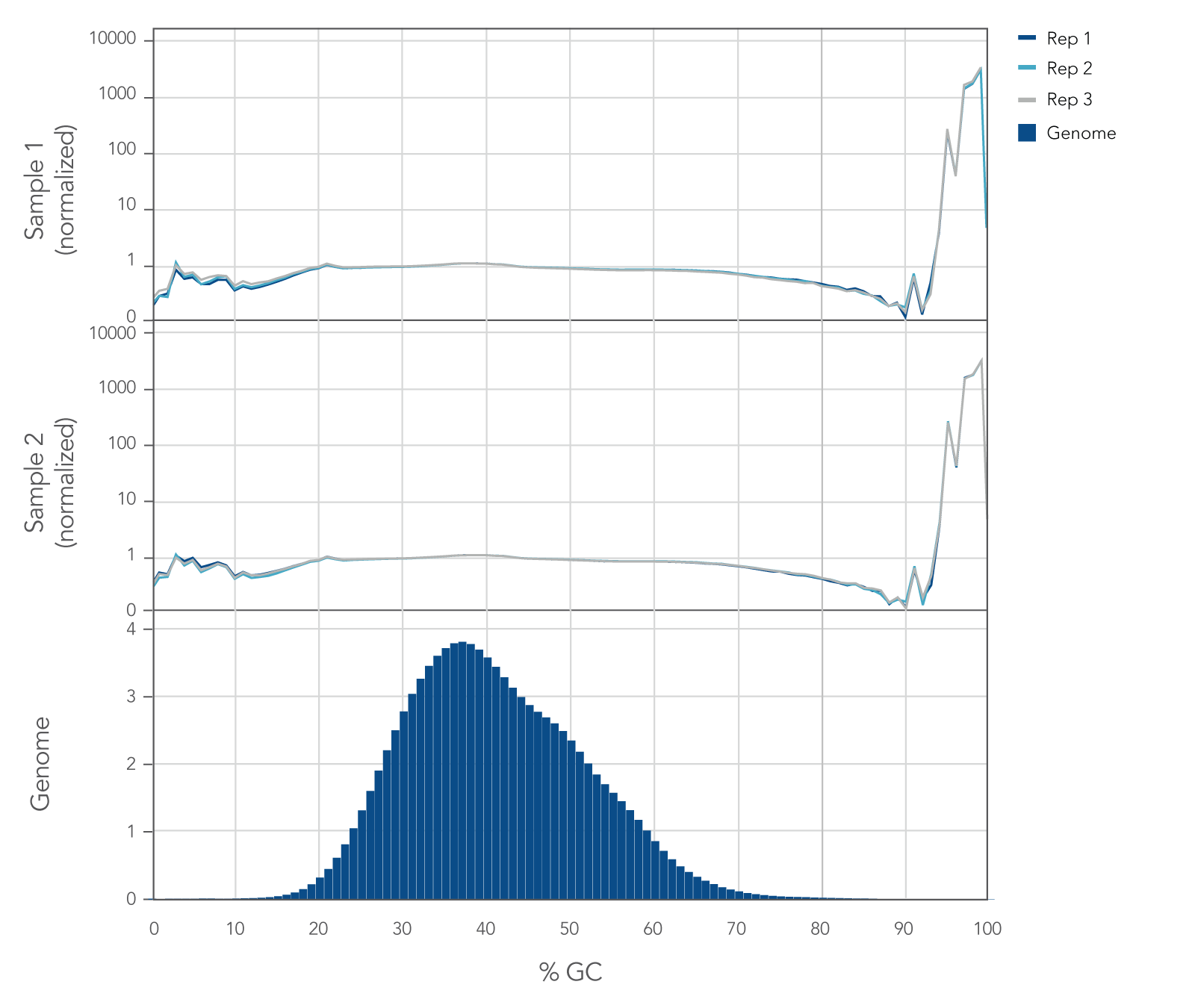
Internal whole genome sequencing (WGS) experiments demonstrated that the xGen cfDNA & FFPE DNA Library Prep v2 MC Kit has even coverage across the human genome with little evidence of bias (Figure 5), resulting in no significant GC bias for 99.7% of the human genome. Even coverage is made possible by the reduced number of PCR cycles required, due to higher conversion rates of input molecules.
Figure 5. xGen cfDNA & FFPE DNA Library Prep Kit normalized GC coverage across the human genome from cfDNA. 10ng cfDNA libraries were generated using xGen cfDNA & FFPE DNA Library Prep v2 MC Kit, using cfDNA extracted from two healthy donors from BioChain. Libraries were generated in triplicate using the manufacturer’s protocol, with 11 cycles of PCR. Libraries were quantified with Qubit dsDNA HS Kit and average size was determined with the Tapestation 4200 High Sensitivity D1000 (Agilent). Libraries were then pooled equimolar and sequenced on a Nextseq 500 (Illumina), using a high-output 300 cycle kit following the manufacturer’s protocol. The cfDNA libraries were subsampled to 20 million paired end reads for analysis with <0.2% of the genome bin at <15% GC and <0.1% of the genome bin at >80% GC.
Resources
Frequently asked questions
What is the new name of the xGen™ Prism™ DNA Library Prep Kit?
The xGen Prism DNA Library Prep Kit has been renamed to the xGen cfDNA & FFPE DNA Library Prep MC v2 Kit.
Should the xGen™ ssDNA & Low-input DNA Library Prep Kit be used for FFPE samples?
In most cases using FFPE samples, we would recommend using the xGen cfDNA & FFPE DNA Library Prep Kit.
In scenarios with heavily nicked or denatured DNA resulting from decrosslinking or other high temperature steps, the xGen ssDNA & Low-Input DNA Library Prep Kit can yield higher recovery and conversion rates of input DNA.
Do I need to titrate the adapters when using different input amounts with the xGen™ cfDNA & FFPE Library Prep MC v2 Kit ?
What is the best quantitation method for assessing the quality and quantity of my DNA before preparing libraries with the xGen™ cfDNA & FFPE Library Prep MC v2 Kit?
We recommend using fluorescent-based DNA quantification tools such as the Qubit™ Fluorometer (Thermo Fisher Scientific) to determine the concentration of your DNA sample. Digital electrophoresis (e.g., Bioanalyzer™ instrument, Agilent) can be used to assess the input sample size distribution.
For formalin-fixed, paraffin-embedded (FFPE) samples, we recommend using standard quality control methods, including finding the Q-ratio with qPCR or the DNA Integrity Number (DIN) using size distribution (e.g., Bioanalyzer instrument). These methods can help you choose the appropriate number of PCR cycles for your sample.
For cell-free DNA (cfDNA), we recommend assessing the size distribution with electrophoresis.
Note, if large molecular weight DNA is present, an additional cleanup may be necessary to remove genomic DNA contamination.
Which adapters should I use with the xGen™ cfDNA & FFPE Library Prep MC v2 Kit?
The Ligation 1 and Ligation 2 Adapters provided in the xGen™ cfDNA & FFPE Library Prep MC v2 Kit should be used for library construction.
Since these adapters attach partial adapter sequences, library amplification using xGen UDI Primer Pairs is required to incorporate sample indexing sequences and add the P5 and P7 sequences.
Which platform can be used for sequencing libraries made with the xGen™ cfDNA & FFPE Library Prep MC v2 Kit?
Why does the xGen™ cfDNA & FFPE Library Prep MC v2 Kit have fixed instead of random UMIs?
Fixed UMI sequences help to enable identification and correction of sequencing or PCR errors, even if they appear within the UMI sequence.
During error correction, using fixed UMI sequences reduces the chance that an error in UMI sequences will accidently be counted as a separate original molecule.
Are there stopping points in the xGen™ cfDNA & FFPE DNA Library Prep MC v2 protocol?
Additionally, there is a pause point after the ligation 1 step, where the reactions can be kept at 4°C for up to 2 hours.
What are the best applications for the xGen™ cfDNA & FFPE Library Prep MC v2 Kit?
The xGen™ cfDNA & FFPE Library Prep MC v2 Kit is designed to help improve variant calling from damaged or degraded DNA samples like cell-free DNA (cfDNA) and DNA extracted from formalin-fixed, paraffin-embedded (FFPE) samples.
How do I check the quality of sequencing libraries made with the xGen™ cfDNA & FFPE Library Prep MC v2 Kit?
Do I need to shear or fragment my DNA before using the xGen™ cfDNA & FFPE Library Prep MC v2 Kit?
DNA has to be an appropriate size before library construction with the xGen™ cfDNA & FFPE Library Prep MC v2 Kit.
For genomic DNA or DNA derived from formalin-fixed, paraffin-embedded (FFPE) samples, we recommend using Covaris shearing to attain average insert sizes of 150–300 base pairs.
As cell-free DNA (cfDNA) typically has an average size of 160 base pairs, no further fragmentation is required.
How do I use UMIs for error correction?
Depending on your sample type or experiment goals, you can choose to use UMIs or ignore them altogether.
The xGen™ cfDNA & FFPE Library Prep Kit Analysis Guidelines leads you through the recommended analysis pipeline using open-source tools starting with FASTQ files and resulting in variant calling.
As an overview, fixed UMI sequences, such as those used with the xGen cfDNA & FFPE Library Prep Kit, enable identification and correction of sequencing or PCR errors, even if they appear within the UMI sequence.
- Single-read families analysis: UMIs can also be used to correct errors in sequencing data at the same time as removing duplicate reads. For example, all reads with the same start-stop position and UMI can be grouped as a single-read family then collapsed. Rather than simply choosing the highest quality read, this method uses all reads within the single-read family to choose the most likely base at each position from beginning to end. This process yields a collapsed single-read family that can be used for variant calling. This approach is taken by the tools GroupReadsByUmi plus CallMolecularConsensusReads (fgbio).
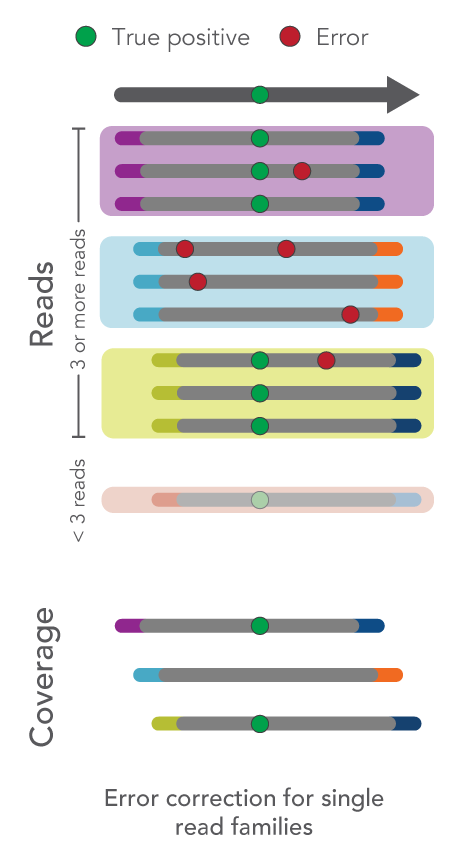
Figure 1. Schematic of error correction methods with UMIs.