Choose your region, country/territory, and preferred language
xGen™ Acute Myeloid Leukemia (AML) Cancer Hybridization Panel
Predesigned panel that targets AML-associated genes
The xGen Acute Myeloid Leukemia (AML) Cancer Hyb Panel comprises 11,731 xGen Hyb Probes, spanning 1.19 Mb of the human genome, for research using targeted enrichment of >260 AML-associated genes, for more efficient identification of disease-causing mutations in research samples.
xGen NGS—made for acute myeloid leukemia research.
Ordering
- 98% of genomic targets covered at greater than 0.2X mean coverage
- Detect variations
- Empirically derived targets
- Enjoy fast turnaround via easy online ordering and next day shipping
Transform Your NGS Workflow with Automation
Looking to streamline your NGS workflows? Discover how automation can enhance efficiency and consistency in your lab with our NGS Automation solutions.
For the xGen Hybridization and Wash Kit, xGen Universal Blockers, xGen Library Amplification Primer Mix, and/or xGen Human Cot DNA, please visit the xGen Hybridization Capture Core Reagents page.
Product details
The xGen AML Cancer Hyb Panel is composed of individually synthesized and quality controlled xGen Hyb Probes that have a 5’ biotin group for capture with streptavidin-coated beads. The panel consists of 11,731 xGen Hyb Probes targeting empirically derived regions within 260 acute myeloid leukemia (AML)-associated genes.
Panel composition (target genes)
Table 1: Frequently researched genes included in the xGen AML Cancer Hyb Panel. *
| ASXL1 | FAM154B | IDH2 | KDR | NPM1 | PRAMEF2 |
| C17orf97 | FAM47A | JAK1 | KIT | NRAS | PTPN11 |
| CBL | FAM5C | JAK2 | KRAS | NTRK3 | RUNX1 |
| CEBPA | FLRT2 | JAK3 | LRRC4 | OR13H1 | TET2 |
| DNMT3A | FLT3 | KCNA4 | MLL3 | OR8B12 | TP53 |
| EGFR | GJB3 | KCNK13 | MYC | P2RY2 | TUBA3C |
| EZH2 | IDH1 | KDM6A | NF1 | PCDHB1 | WT1 |
* The panel does not target every exon in each gene, but targets all regions found to be mutated in the TCGA study [1].
Product data
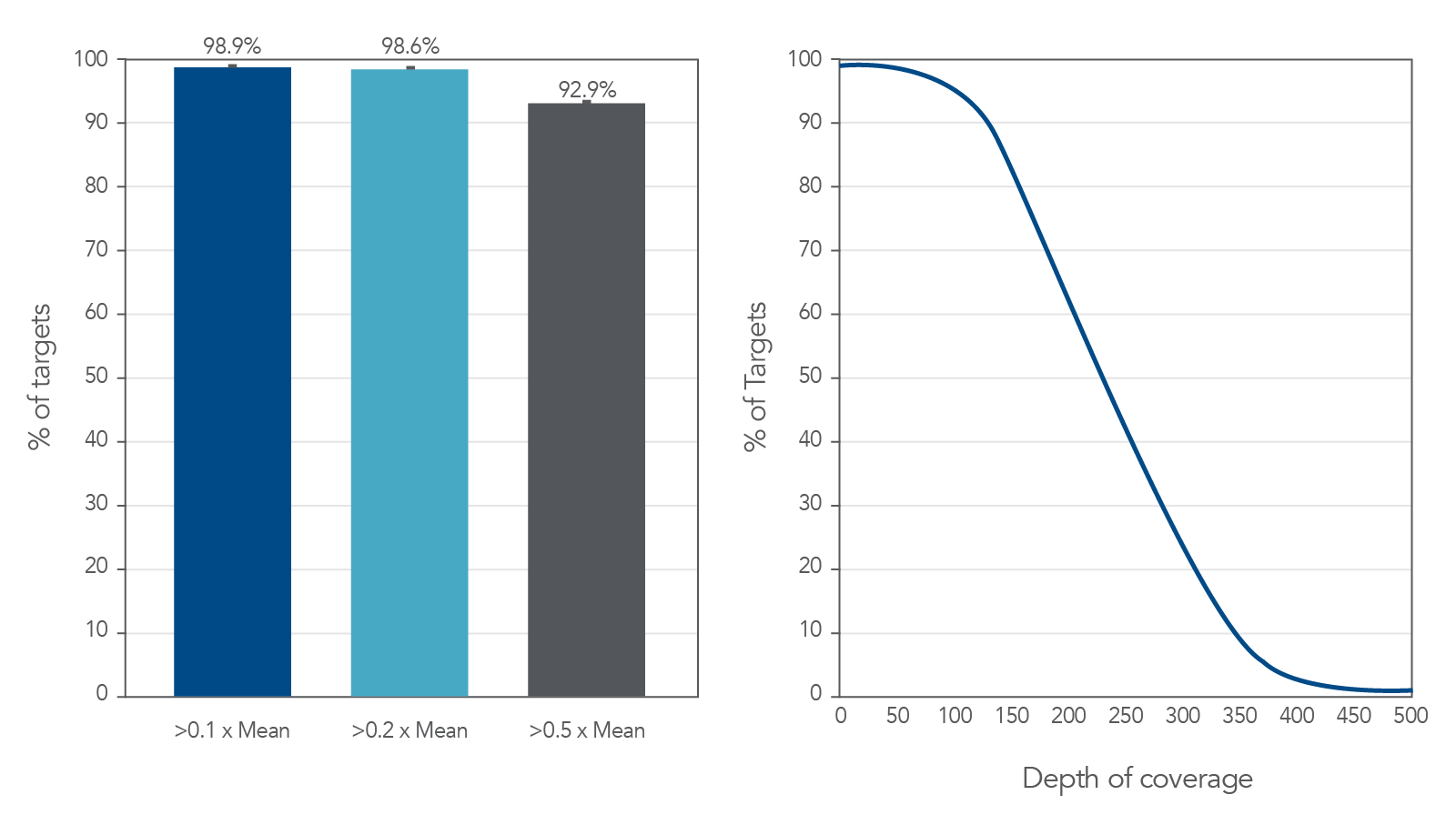
Coverage uniformity for the xGen AML Cancer Hyb Panel
Figure 1. Coverage uniformity obtained with xGen AML Cancer Panel. Greater than 0.2X mean coverage is observed for >98% of targets. Illumina TruSeq® HT libraries (n=8) were enriched using the xGen AML Cancer Panel and sequenced
on a NextSeq 550 system using 2 x 150 paired-end reads. Each sample was sub-sampled to 4M total reads.
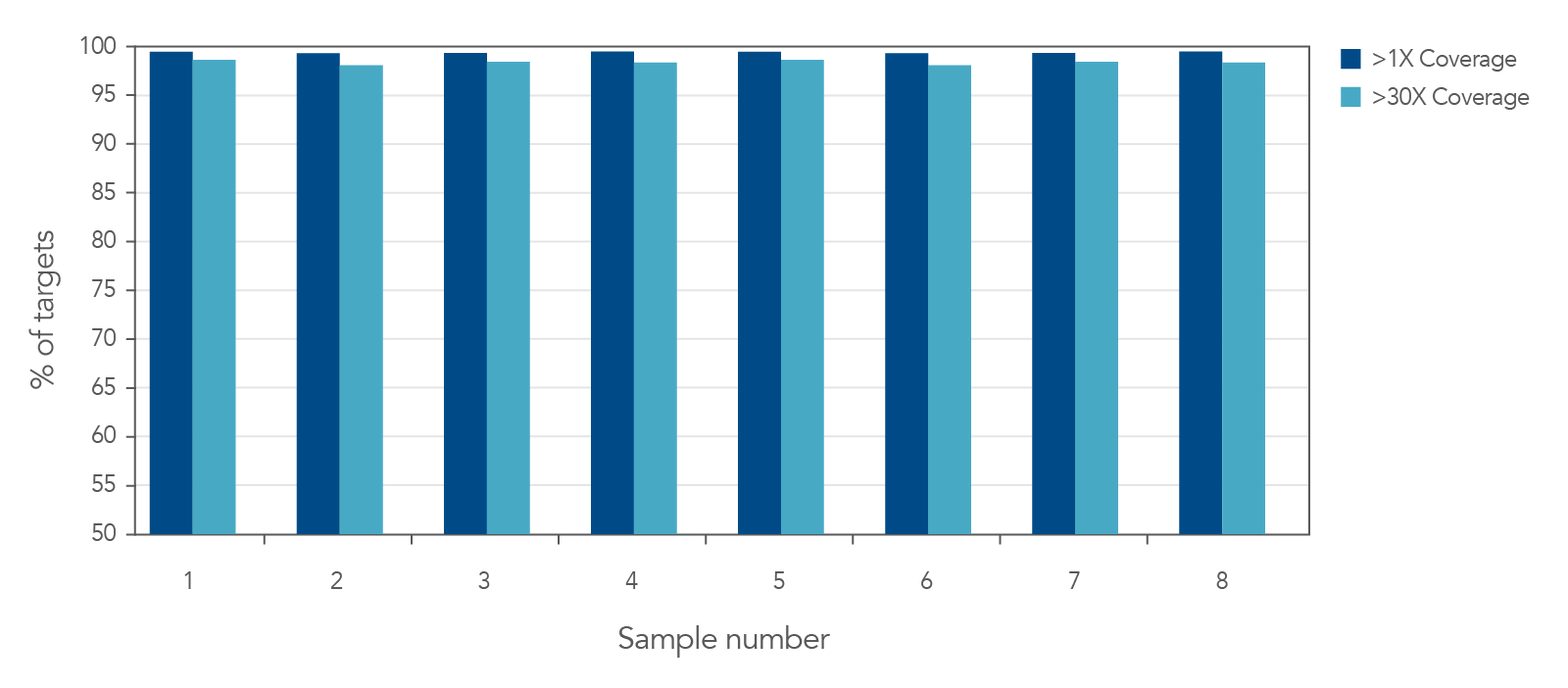
Figure 2. Deep coverage of targeted regions using xGen AML Cancer Panel. Eight genomic DNA libraries were enriched using the xGen Acute Myeloid Leukemia Cancer Panel and sequenced on a NextSeq 550 system using 2 x 150 paired-end reads. Total reads for each sample were sub-sampled to 4M total reads. In all samples, there was >1X coverage for 99.3% of targets and >30X coverage for 98.7% of targets.
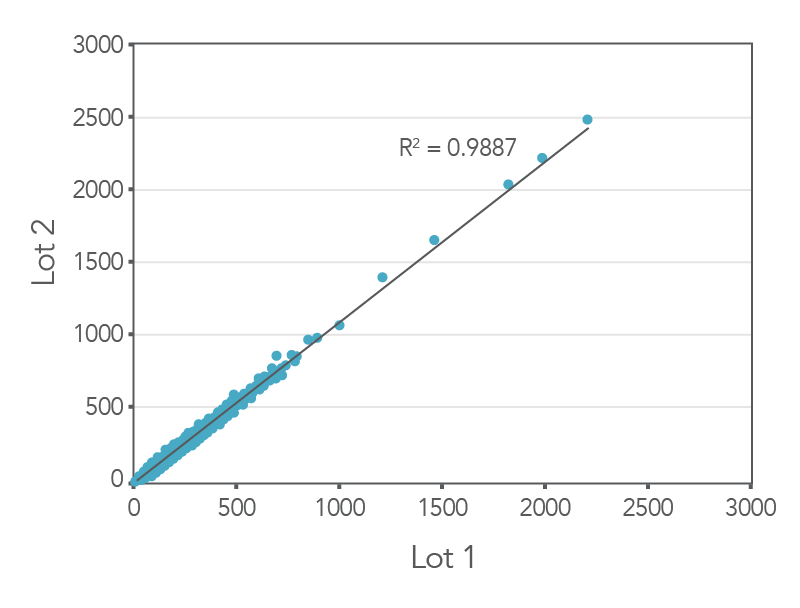
Consistent results between lots
Figure 3. Consistent results obtained with xGen AML Cancer Panel. Genomic DNA libraries were enriched with two lots of the xGen AML Cancer Panel (n=8 for each lot). The enriched samples were sequenced on the NextSeq
550 sequencing platform using 2 x150 paired-end reads. All samples were sub-sampled to a level of 4M total reads. The probe-by-probe target coverage was averaged for eight replicate samples for each lot of the xGen AML Cancer panel. A comparison of
probe-by-probe target coverage between two lots show consistent results between lots, with an R2 value of 0.9887.
Resources
References
- Cancer Genome Atlas Research N, Ley TJ, Miller C, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059-2074.