Choose your region, country/territory, and preferred language
xGen™ Custom NGS Hybridization Capture Panels
Custom target enrichment panels for maximum flexibility
xGen Custom Hyb Panels include your choice of high-fidelity, individually synthesized, 5′-biotinylated oligos for targeted NGS research. IDT proprietary DNA synthesis produces rapid, high-quality panels that can be optimized, expanded, and combined with other panels in research studies.
xGen™ NGS—made custom.
Ordering
xGen™ Custom Hyb Panels
- Includes an NGS Functional Test Report† with specific capture metrics to ensure confidence
- Delivered typically in as few as 3 weeks
- Accel available in 5 business days
- Affordable options available for any size panel
- Create your custom panel design using xGen Panel Design Tool
- Reliable lot-to-lot results instills confidence in panel functionality*
- Production provides additional quality documentation, including ESI/MS on individual probes
†Not provided with the xGen Custom Hyb Panel—Accel option
*As shown in figure below
xGen Custom Hyb Panels have been optimized for use with the xGen Hybridization and Wash kit, xGen Universal Blockers, and xGen Library Amplification Primer Mix, which can be ordered from the xGen Hybridization Capture Core reagents page.
Create designs for custom hyb panels
For designing custom hyb panels, please use the xGen Hyb Panel Design Tool.
Use xGen Hyb Panel design toolOrder my design
If you have predesigned probe sequences, upload them using our submission form.
Order my designRequest design consultation
For more information on xGen Custom Hyb Panels, please contact IDT Sales
Request design consultationProduct details
The IDT xGen Custom Hyb Panels consist of high-fidelity, individually synthesized, 5′-biotinylated oligos. Each custom panel is created by mixing equimolar concentrations of each probe, which promotes equal representation of each probe in the final product.
IDT offers a custom design pipeline with technical support in xGen Custom Hyb Panels to answer any of your questions about our custom services. The IDT NGS Functional Test Report is provided with the standard xGen Custom Hyb Panel and the xGen Custom Hyb Panel—Production, which provides evidence that you are receiving a working panel.
The xGen Custom Hyb Panel is delivered typically within 3 weeks of placing your order. For those users who would like to receive their panels sooner, the IDT xGen Custom Hyb Panel—Accel can be delivered in as few as 5 business days. To meet the accelerated delivery, the xGen Custom Hyb Panel—Accel does not include NGS functional testing. Finally, the xGen Custom Hyb Panel—Production and xGen Custom Hyb Panel—Production (No functional testing) options are for customers who need quality control metrics of their panel’s manufacturing through additional quality documentation, including individual probe ESI/MS.
Overall, IDT can provide a solution that fits any size project. Whether it needs more design iteration, larger reaction sizes, or more quality documentation, we offer customized products that fit your research needs.
Looking to streamline your NGS workflows? Discover how automation can enhance efficiency and consistency in your lab with our NGS Automation solutions.
| Feature | xGen Custom Hyb Panel | xGen Custom Hyb Panel—Accel | xGen Custom Hyb Panel—Production | xGen Custom Hyb Panel—Production (No functional testing) |
|---|---|---|---|---|
| Custom design service | ✓ | ✓ | ✓ | ✓ |
| Quality synthesis | ✓ | ✓ | ✓ | ✓ |
| NGS Functional Test Report | ✓ | ✓ | ||
| Quality documentation | ✓ | ✓ | ||
| Stock custom panel | ✓ | ✓ | ✓ | ✓ |
| Reaction sizes | 16, 96, custom | 16, 96, custom | 16, 96, custom | 16, 96, custom |
| Turnaround time | Typically within 3 weeks | Within 5 business days | Project dependent | Project dependent |
Product data
The IDT xGen Custom Hyb Panels are screened for off-target hybridizations with a proprietary new quality control (QC) method. IDT bioinformatics scientists have created the xGen Off-Target QC Method that uses a sequence identity-based approach to screen your custom hybridization probes. This method can expand the size of your target space without increasing the risk of off-target capture (Figure 1).
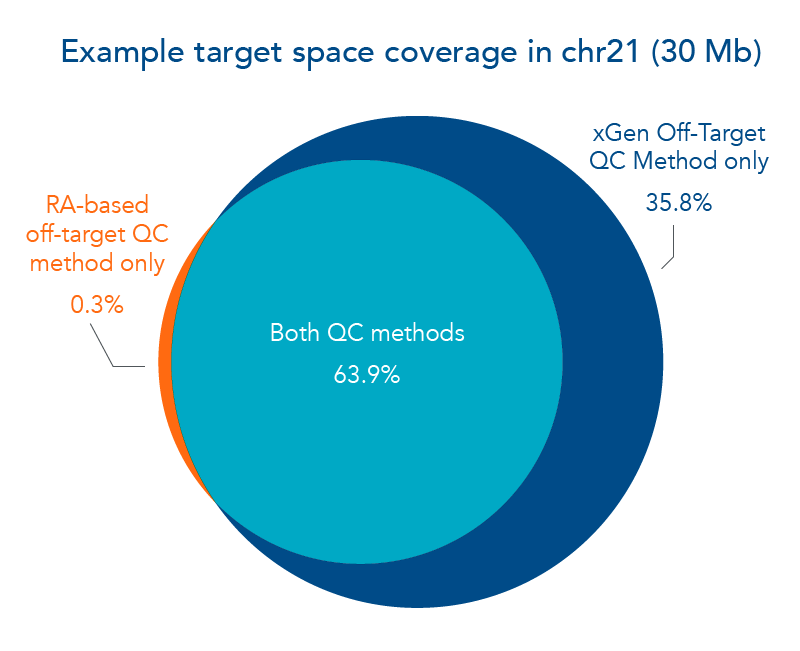
Figure 1. The xGen Off-Target QC Method expands target region. In this example, a panel of hybridization probes created to capture fragments of human chromosome 21 were screened for off-target hybridization using 2 different QC methods. The repeat annotation (RA) method was compared to the xGen Off-Target QC Method in their ability to refine a custom hybridization panel that maximized the target region without increasing the amount of off-target hits. As shown in the Venn diagram, 63.9% of the example target space passed both off-target QC methods. Notably, 35.8% of the example target space passed the xGen Off-Target QC Method, whereas 0.3% of the example target space passed only the RA-based off-target QC method.
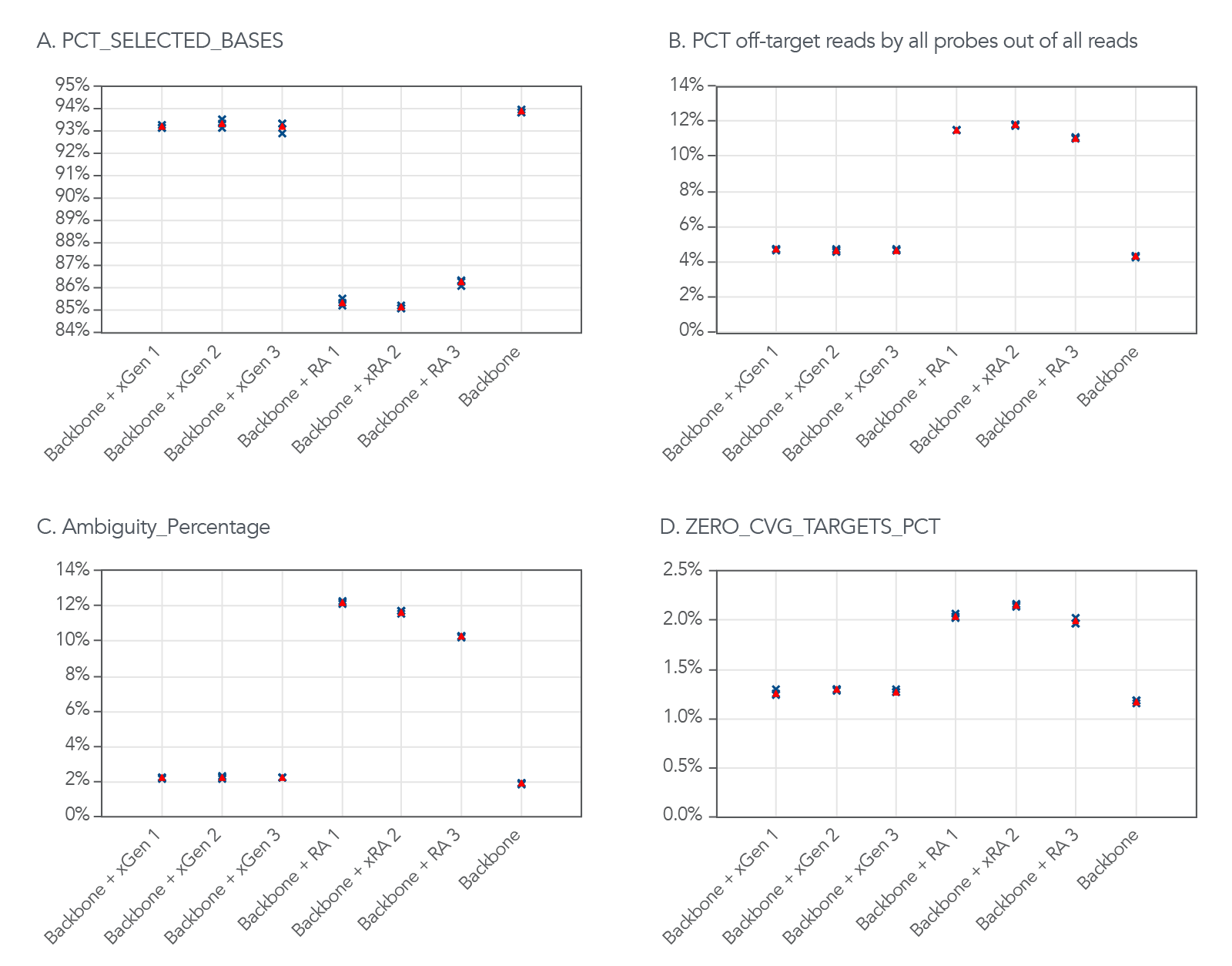
Although the xGen Off-Target QC Method expanded the target region of this example panel, the panel still maintained a high on-target rate. Using the Picard HsMetrics algorithm (Broad Institute), the standard on-target rate metric (PCT_SELECTED_BASES) was used to investigate the on-target rate. In 3 different experiments, using both QC methods, the xGen Off-Target QC Method had a higher percentage of the targeted bases covered (Figure 2A) and a lower percentage of off-target reads (Figure 2B). In addition, the percentage of ambiguity and zero coverage targets were lower (Figure 2C–D).
Figure 2. The xGen Off-Target QC Method does not increase the amount of off-target reads. (A) The xGen Off-Target QC Method adds design space with minimal impact on the on-target rate compared to the repeat annotation based off-target QC method. (B) The xGen Off-Target QC Method minimizes the number of off-target reads contributed by the additional probes compared to the repeat annotation based off-target QC method (C-D). The xGen Off-Target QC Method adds additional design space without increasing the percentage of ambiguity and zero coverage targets. The blue X’s represent experimental replicates, and the red triangles represent the average of three replicates.
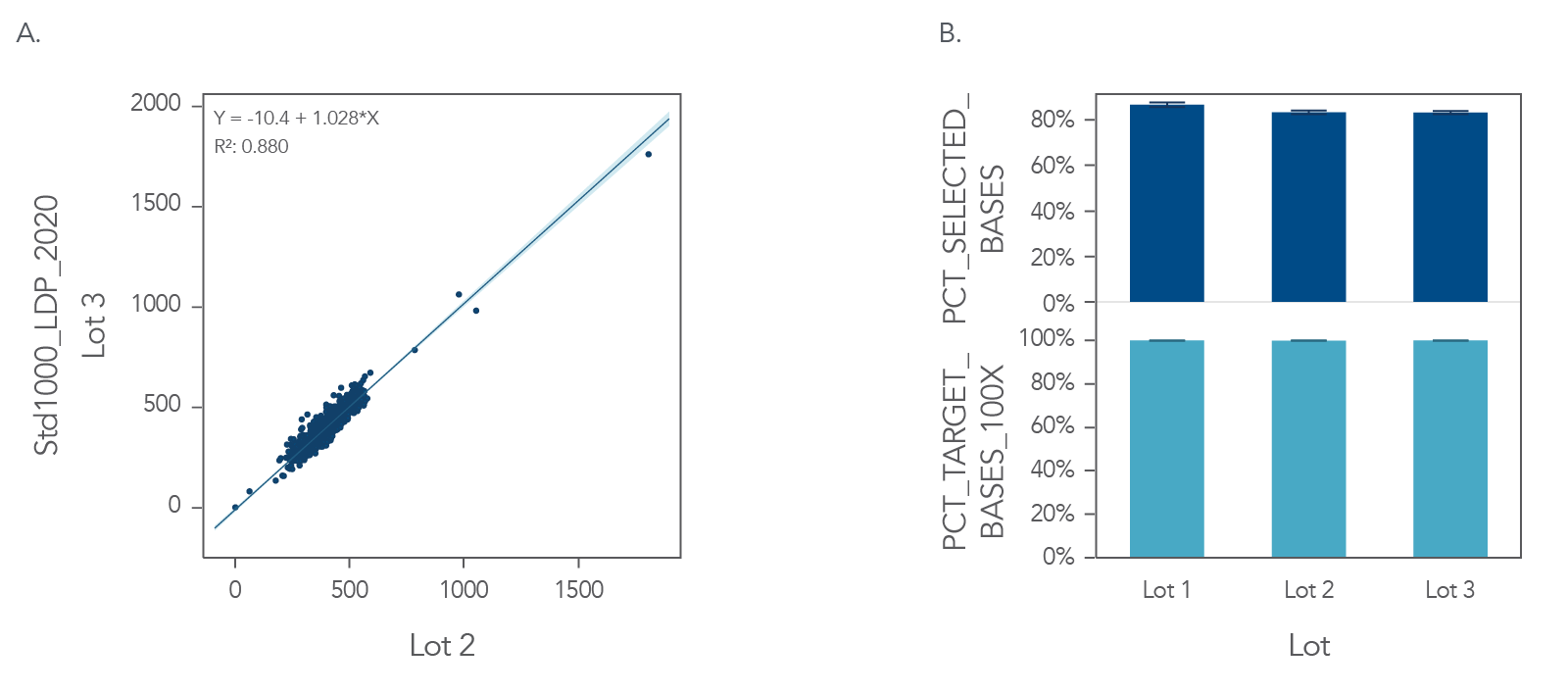
Figure 3. Example of uniform sequence coverage and high on-target rates with xGen Custom Hyb Panels. DNA libraries created from 100ng of one (1) human genomic DNA (Coriell NA12878), in triplicate, run on 3 lots on 1 day and were enriched for an 89 kb target region, using an xGen Custom Hyb Panel—Production. The enriched libraries were sequenced on an Illumina NextSeq® 550 instrument and subsampled to 1M reads. The data show (A) consistent mean target coverage for 2 synthesis lots of 999 probes (a subset of the full experimental data shown here). The mean of triplicates for each synthesis lot are plotted. On-target rates (top) and coverage (bottom) for 3 synthesis lots were calculated with Picard metrics (Broad Institute), and plotted as mean of triplicates (B).
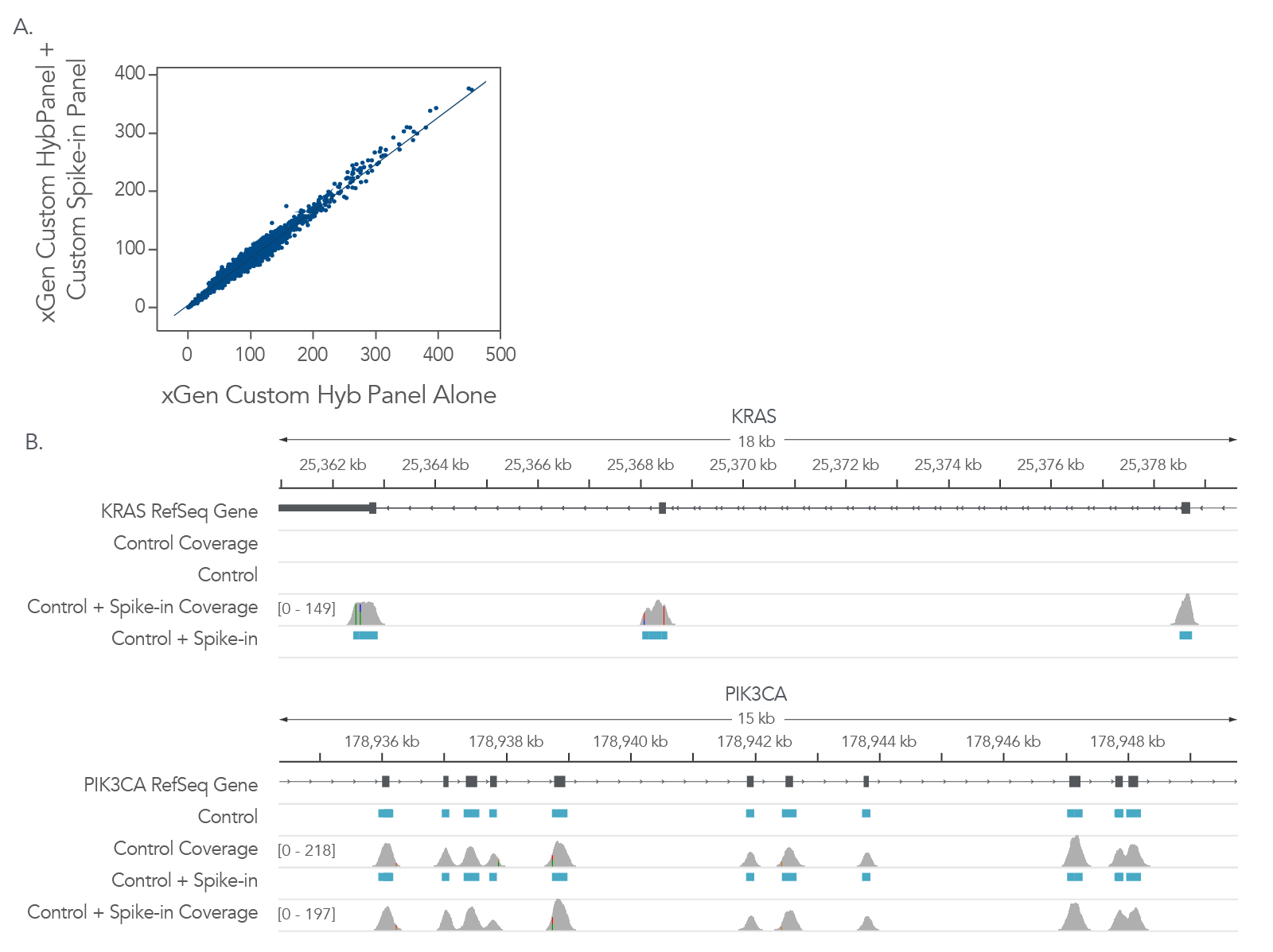
Often, there is a need for expanding an existing panel to accommodate new information about your research interests. IDT xGen Custom Hyb Panels can be expanded by adding xGen Custom Hyb Probes to the new targets. As shown in Figure 4, the addition of Custom Hyb Probes in this example, did not impact the coverage of the original panel. Using panels constructed from individually synthesized probes is key to the flexibility. Mixing individual probes into the final panel ensures that each probe is represented, and the molar concentrations are relatively equal for each probe in the panel. The additional probes can be added to the existing panel at the correct molar concentration.
Figure 4. xGen Custom Hyb Probes can be added to xGen Custom Hyb Panels to expand the target regions of interest (ROIs). To customize an existing hyb panel, xGen Custom Hyb Probes can be added to the existing panels. Libraries were prepared from 100ng of one (1) human genomic DNA sample (Coriell NA12878), in triplicate, on one (1) day and captured using either an xGen Custom Hyb Panel (5001 probes targeting 33 genes, covering a 600 kb region) or the same panel spiked with additional xGen Custom Hyb Probes targeting 32 additional genes (89 kb). (A) Coverage of xGen Custom Hyb Panel, with and without the spike-in of xGen Custom Hyb Probes, demonstrates that adding additional probes does not impact coverage of the main panel. (B) Representative Integrated Genome Viewer (IGV) screenshots are shown for PIK3CA (targeted in main xGen Custom Hyb Panel) and KRAS (additional content from spike-in xGen Custom Hyb Probes). Probe regions are indicated in blue.
To make sure that you receive a working hybridization panel, the IDT xGen Custom Hyb Panels and xGen Custom Hyb Panels—Production include functional testing prior to customer delivery. Your panel is used to prepare NGS libraries from 100ng Coriell genomic DNA (gDNA) (2 samples in triplicate). Each NGS Functional Test Report includes statistics of the final sequencing data after 2 x 150 bp paired-end runs on an Illumina® sequencing instrument. Using Picard suite of analysis tools (Broad Institute), the report includes key metrics such as total sampled reads, mean target coverage, percent selected bases, percent duplication, and AT and GC dropouts, among others.
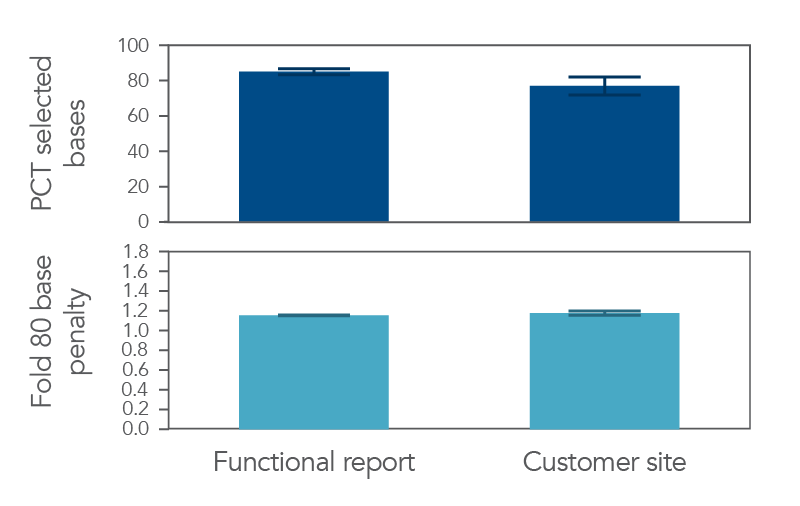
Data obtained from the panel by the customer emulated the data presented in the IDT NGS Functional Test Report (Figure 5), thereby demonstrating that the NGS Functional Testing summary provides you with the confidence that your xGen Custom Hyb Panel will perform as expected.
Figure 5. xGen Custom Hyb Panels are functionally tested prior to delivery. 100 ng of two (2) human genomic DNA samples (Coriell NA12878 and NA24385) sample was used to prepare NGS libraries. 500 ng of these libraries (n=5) were manually captured in triplicate, on one (1) day with a custom lung cancer panel (208 kb). Data from this experiment were used to generate a functional NGS report (left). The same custom lung cancer panel was then captured by the customer on a Biomek i7 (n=14, right). NGS functional data is pulled from Picard HS Metrics. All samples achieved 100X coverage or deeper.
Resources
Frequently asked questions
What are the new names of IDT next generation sequencing products?
After IDT acquired Swift Biosciences, some of the IDT next generation sequencing products were renamed.
The following table lists the previous IDT product name corresponding with its new name, hyperlinked to the appropriate product page.
| Previous IDT product name | New IDT product name |
|---|---|
| xGen™ Prism™ DNA Library Prep Kit | xGen cfDNA & FFPE DNA Library Prep MC |
| COVID-19 Capture Bundle | xGen SARS-CoV-2 Hyb Panel Bundle |
| xGen CNV Backbone Panel—Tech Access | xGen CNV Backbone Hyb Panel |
| xGen Human ID Research Panel | xGen Human ID Hyb Panel |
| xGen Human mtDNA Research Panel v1.0 | xGen Human mtDNA Hyb Panel |
| xGen Exome Research Panel v2 | xGen Exome Hyb Panel v2 |
| Lotus™ DNA Library Prep Kit | xGen DNA Library Prep Kit MC or xGen DNA Library Prep Kit EZ |
| Custom or predesigned rhAmpSeq™ Library Prep Kits | Discontinued, but technology is available via rhAmpSeq for CRISPR Analysis System |
| Discovery Pools | xGen Custom Hyb Panels and xGen Custom Hyb Panels—Accel |
| Lockdown™ Probes Functional QC Discovery Pool | xGen Custom Hyb Panels—Production (available with and without functional testing) |
| xGen Predesigned Gene Capture Pools | xGen Custom Hyb Panels |
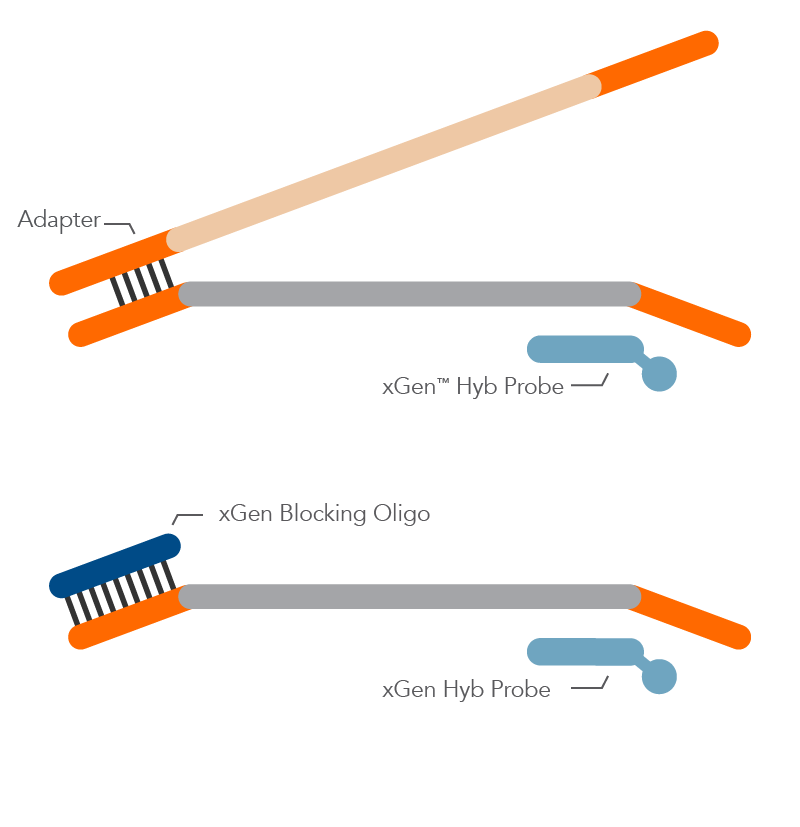
What is the difference between universal and standard blocking oligos (used in NGS experiments)?
Standard blocking oligos are designed for specific barcode sequences. While these designs can be effective in blocking daisy-chaining between fully indexed library molecules, it can be quite cumbersome for the researcher to manage individual blocking oligos that mate with specific indexes, as the researcher expands the number of samples to multiplex together.
The xGen™ Universal Blocking Oligos solve this problem of managing a 1:1 blocker to adapter matching by using a proprietary oligo design to allow our universal blockers to block effectively across ALL index designs of a particular length, while also boosting the on-target percentage rate as compared to standard blocking oligos.
How do I determine the number and cost of target capture probes needed to target N genes/gene regions on my xGen™ Custom Hyb Panel?
You should consider target enrichment (target capture) if you are analyzing a subsection of a genome or performing deep sequencing to identify rare mutations.
This helps you to reserve sequencing capacity for the region(s) of interest and reduce the sequencing cost and time associated with data analysis.
Our xGen™ Custom Hyb Panel probes are practical because they can be used on their own, or for supplementing other capture panels to expand the target region and improve enrichment.
Can you make xGen™ Custom Hyb Panel probes based on our design results from another vendor?
Yes.
Generally speaking, all IDT needs are the capture probe sequences to synthesize xGen™ NGS Hybridization Capture probes.
For reference, IDT uses NCBI databases.