CRISPR genome editing can be used to modify genomes by introducing single- or double-stranded breaks (DSB) which are then repaired using native molecular pathways. When edits are introduced through DNA repair pathways, they can be characterized using targeted next generation sequencing (NGS). Accurate quantification of editing at both on- and off-target sites is paramount to developing applications of CRISPR. To enable easily accessible, accurate analysis of NGS data derived from CRISPR experiments, Integrated DNA Technologies, Inc. (IDT) has created and launched their own cloud-hosted software tool, CRISPAltRations.
Our methodology
CRISPAltRations is a software tool that is accessed through a web interface, rhAmpSeq CRISPR Data Analysis Tool. The tool utilizes cloud-hosted computational resources for data processing. Briefly, this is the workflow:
- The tool identifies and merges read pairs from paired-end sequencing.
- The reads are binned to the expected amplicons resulting from targeted amplification library preparation (e.g., rhAmpSeq CRISPR Library Kit).
- The alignment of the read to the expected amplicon is refined using a Cas-enzyme specific aligner.
- Variants are called and summarized.
Although these steps are relatively common to most software tools that analyze NGS data derived from CRISPR screens, CRISPAltRations has a number of improvements that enable higher accuracy of variant identification, including:
- a Cas-specific aligner
- a specific default variant identification window
- systematic program parameters for open-source tools that will provide quality results
Tool validation
To better understand the impact of these improvements, we developed a set of synthetic datasets to assess the accuracy of annotating on- and off-target editing (11 on-target sites; 592 off-target sites), as well as the accuracy of annotating mutations introduced through the homology directed repair (HDR) pathway at on-target loci (91 on-target sites). The reliability of CRISPAltRations was compared to other published software tools, such as Amplican [1] and CRISPResso2 [2]. For on/off-target characterization, CRISPAltRations characterized the percent of indels down to <0.1% deviation from expectation for 99.5% of target sites. Alternative workflows such as Amplican/CRISPResso2 could not reach this level of precision even with a higher threshold for error (<2% deviation) (Figure 1). 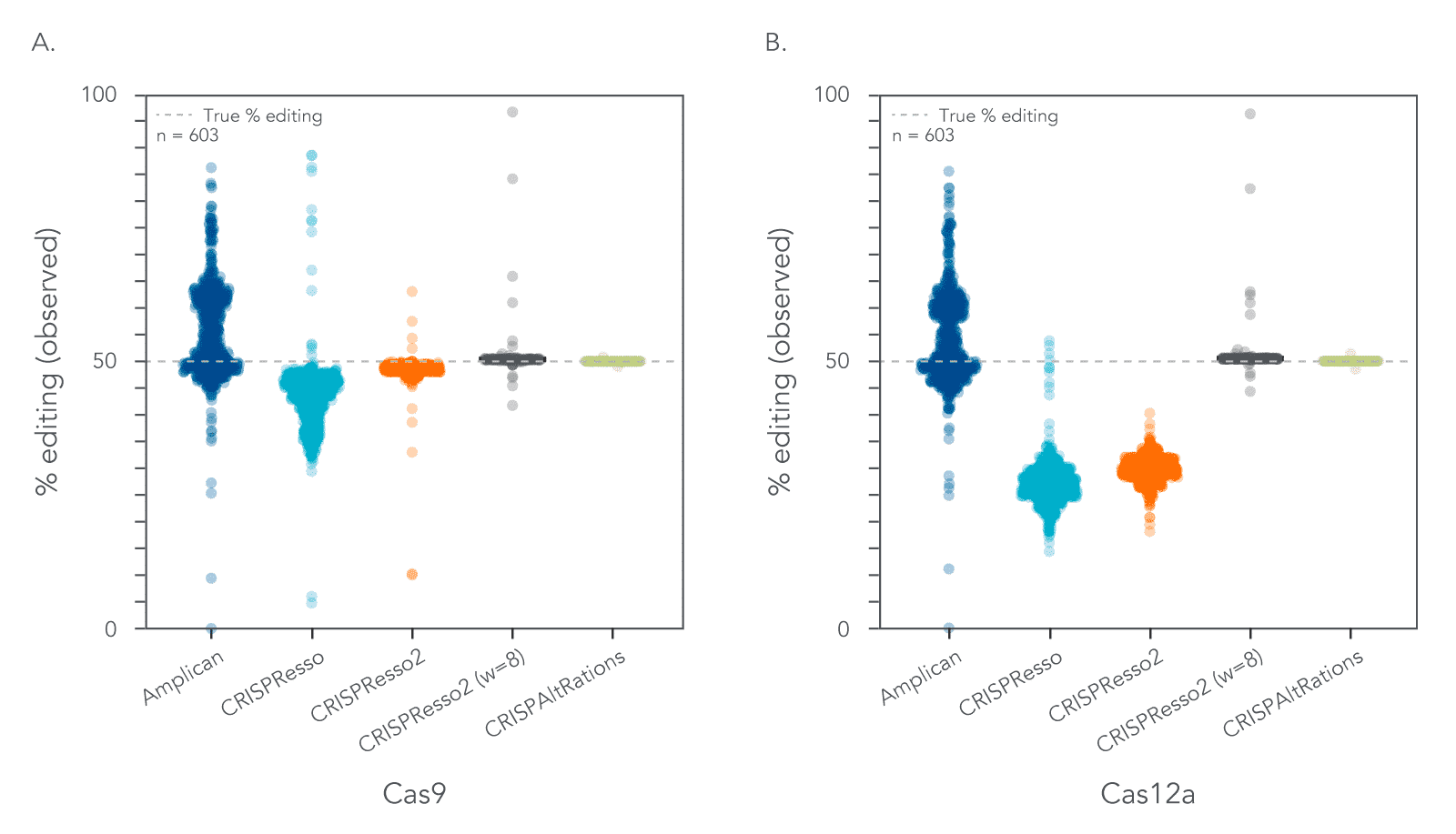
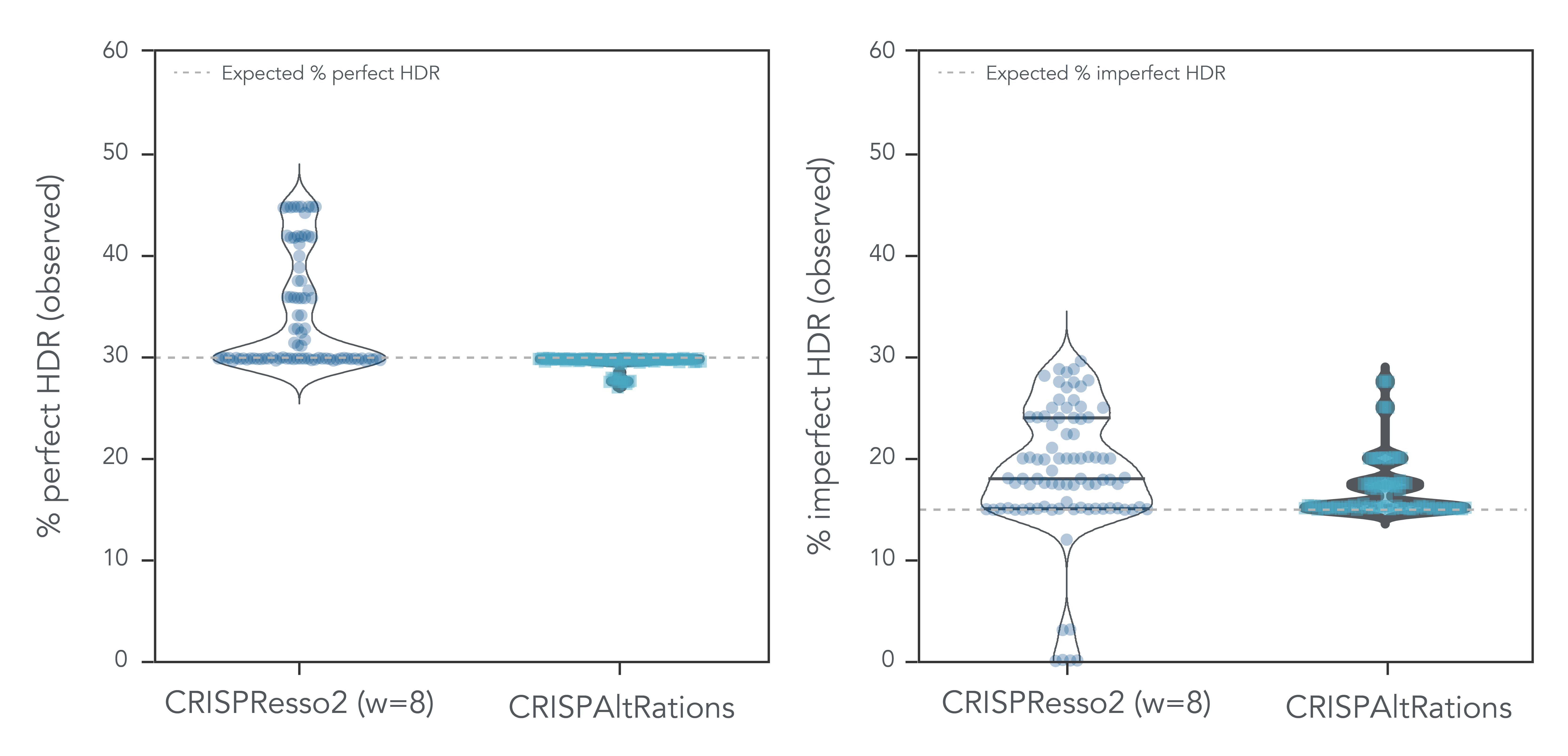
For on-target HDR repair characterization, we compared the quality of CRISPAltRations and CRISPResso2. CRISPResso2 was unable to complete analysis at 4.3% of targets and overestimated the perfect HDR repair events by >3% at 38% of targets (Figure 2). CRISPAltRations provides accurate precision on this dataset with <2% deviation from the expected percent perfect HDR events. Furthermore, CRISPRAltRations can better differentiate an editing event as being derived from the HDR (imperfect) vs NHEJ pathway, as compared to CRISPResso2 (Figure 2).
Experimental recommendations and limits
To further guide experimental design, we generated a series of recommendations for using CRISPAltRations. First, we investigated the read depth requirements to accurately annotate editing at different levels of sensitivity. To do this, we subsampled a series of rhAmpSeq panels with various amounts of on- and off-target editing to compare the annotated indels of subsampled samples to that of the original sample. Generally, there is an inverse correlation between editing efficiency and the number of reads needed for quantification. With our tool, editing annotation can reach ~0.5% editing with only 1000 reads per target (Figure 3). With increased read depth and subtraction of background signal in an unedited control, editing annotation can reach ~0.1% in ideal scenarios. However, background indel noise depends on several factors such as sequence context, sequencer run, library preparation, and more. Thus, low levels of editing should be accompanied by an appropriate statistical test or other advanced methods to ensure confidence of genome editing. Our recommendations are based on using the rhAmpSeq Library Kit followed by 2 x 150 sequencing on a MiSeq™ (Illumina) with v3 chemistry.

Accessibility
To make CRISPAltRations more broadly accessible, we developed a web user interface (UI) that utilizes cloud resources for data processing and storage (Figure 4). This ensures that researchers are not restricted by two major burdens often encountered: 1) a lack of programming knowledge and/or bioinformatics personnel, or 2) a lack of suitable computational resources. This interface enables users to upload data by streaming from local hardware, streaming from cloud resources (AWS/Google/BaseSpace), or simply “dragging-and-dropping” from the web interface. The interface can run thousands of samples simultaneously and includes interactive visualization of the generated results. Advantageously, researchers can easily export results to commonly-used programs (e.g., Excel) to enable customized integration into other existing graphing software to meet specific needs. By providing a version-controlled, tested, and easily-accessible analysis software, we hope to empower the scientific community to use NGS to evaluate the effects of on- and off-target editing resulting from their CRISPR genome editing experiments. 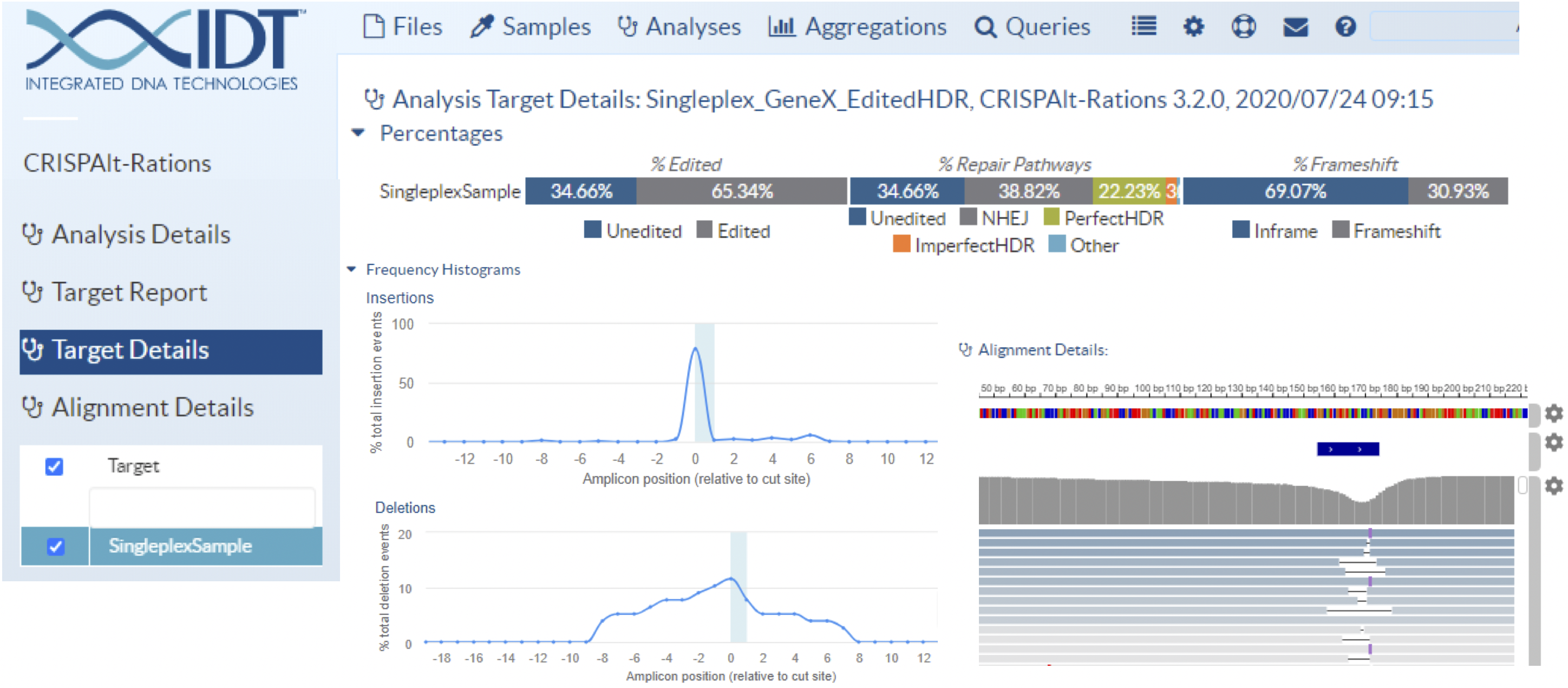
Conclusion
We have developed and deployed a software analysis tool, CRISPAltRations, for accurate quantification of genome editing from CRISPR experiments. We show that a combination of novel features, pin-pointed parameters, and systematically tested code enables us to improve our ability to annotate editing from DSB events. By comparing to other CRISPR analysis tools, we show that CRISPAltRations outperforms other tools for characterization of NHEJ and HDR activity at on/off-target locations. By providing valuable experimental recommendations and developing a “point-and-click” interface we hope to set researchers up for success, regardless of scientific background. We are excited to see what you can do with high-quality genome editing data. Check out the rhAmpSeq CRISPR Analysis System, and get started on your CRISPR analysis.
References
- Labun K, Guo X, Chavez A, et al. Accurate analysis of genuine CRISPR editing events with ampliCan. Genome Res. 2019;29:(5)843–847
- Clement K, Rees H, Canver MC, et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol. 2019;37(3):224–226.
For research use only. Not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical diagnostic use. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations. RUO22-1493_001